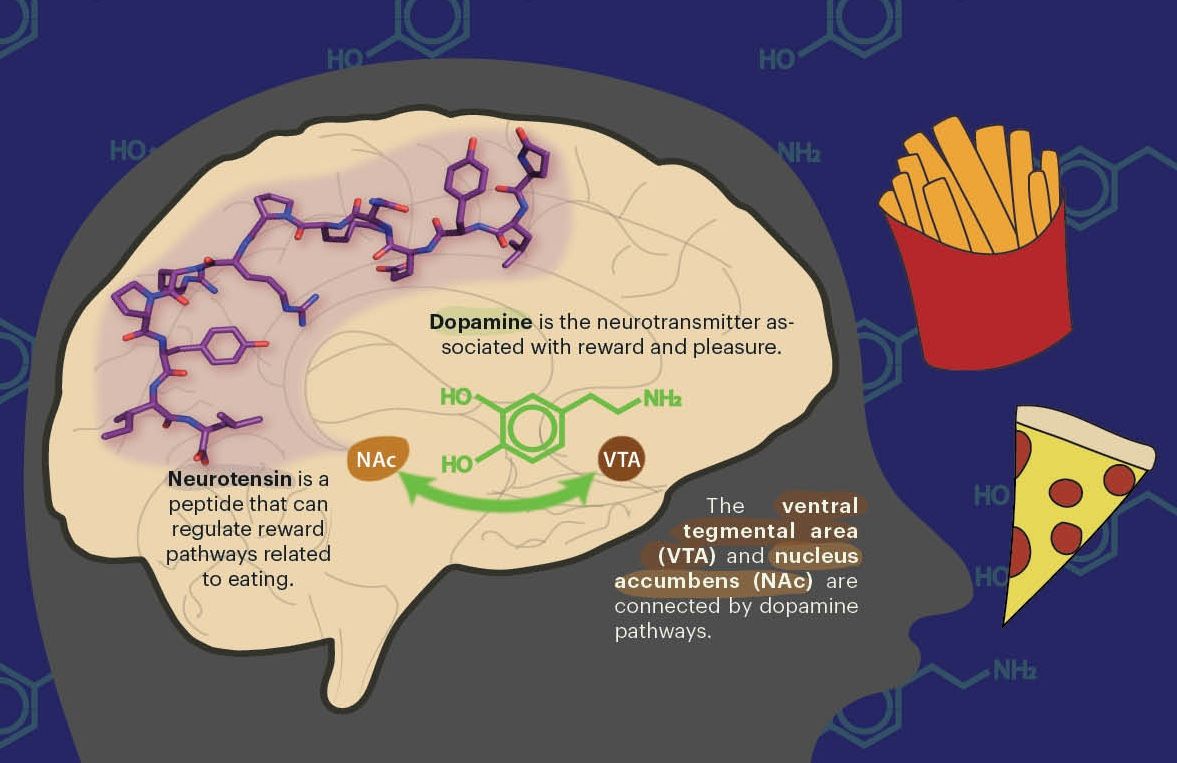
How do you prove something in the past happened? This is the question driving evolutionary biologists like Dr. Marianna Karageorgi, a postdoc working with Professor Noah Whiteman in the Department of Integrative Biology. Karageorgi is fascinated with uncovering the mechanisms of evolution. “It’s one thing to do comparative studies, and say ‘This species is different from this species’ to infer how traits might have evolved,” she notes. “It’s another to say, ‘I’m going to prove this is how it happened.’” One of the holy grails of modern evolutionary biology is figuring out how to test hypotheses about evolution in living organisms. In a recent study, Karageorgi and her collaborators did just that. They engineered specific mutations from the monarch butterfly into the fruit fly, bestowing it the monarch’s resistance to milkweed toxin. In doing so, they bridged two worlds of technologies: statistical methods for developing evolutionary hypotheses, and genome-editing tools for verifying those hypotheses.
Statistics, enabled by lots of data, is the workhorse for evolutionary biologists developing conjectures about an unobservable past. Most evolutionary hypotheses are formulated by sequencing the genes of different species, then using statistical methods to uncover hidden patterns that differ between their DNA. Such patterns can suggest relationships between species, or whether a mutation is associated with a trait of interest, like toxin resistance.
Karageorgi’s team used two statistical methods, phylogenetic inference and ancestral state reconstruction (ASR), to conjecture the gene sequences of the common ancestors of toxin-resistant insects. Species are related to each other through mutations, so the number of mutations between a pair of species indicates how long ago their common ancestor existed. Phylogenetic inference combines this information from every pair of species to reconstruct the tree of relationships between species and their common ancestors, called a phylogeny. At each fork in the tree, which represents a common ancestor, ASR computes the most probable gene sequence, based on inferred rates of mutation and the descendants’ mutations. Together, phylogenetic inference and ASR paint a picture of the evolutionary history of a gene.
However, it’s not always clear which mutations in that history were important. “Most mutations that persist are neutral, meaning they may not affect the function of the gene,” explains Julianne Pelaez, a PhD candidate working with Whiteman. Positions of a gene critical to its function tend to evolve slower, so the team analyzed the rates of evolution inferred from ASR at each position to hypothesize which specific mutations were needed for toxin resistance. They identified two mutations in toxin-resistant insects, occurring at positions 111 and 122 of a particular gene.
Further analysis also flagged a mutation at position 119, which had initially been bypassed because it existed in both toxin-resistant and non-resistant insects. To pinpoint it, Pelaez looked for mutations that co-evolved, or often appeared together, and found that the mutation at 119 always occurred before the one at 122. “If [the mutation] is always present, most likely it has a pre-adaptive role: it’s preventing something bad from happening,” explains Karageorgi. The team estimated how likely this mutational order occurred by chance, using a statistical technique called a permutation test. This probability was low, meaning it was unlikely that 122 always mutated after 119 purely by coincidence. That meant 119 played a critical—if mysterious—role in the evolution of toxin resistance.
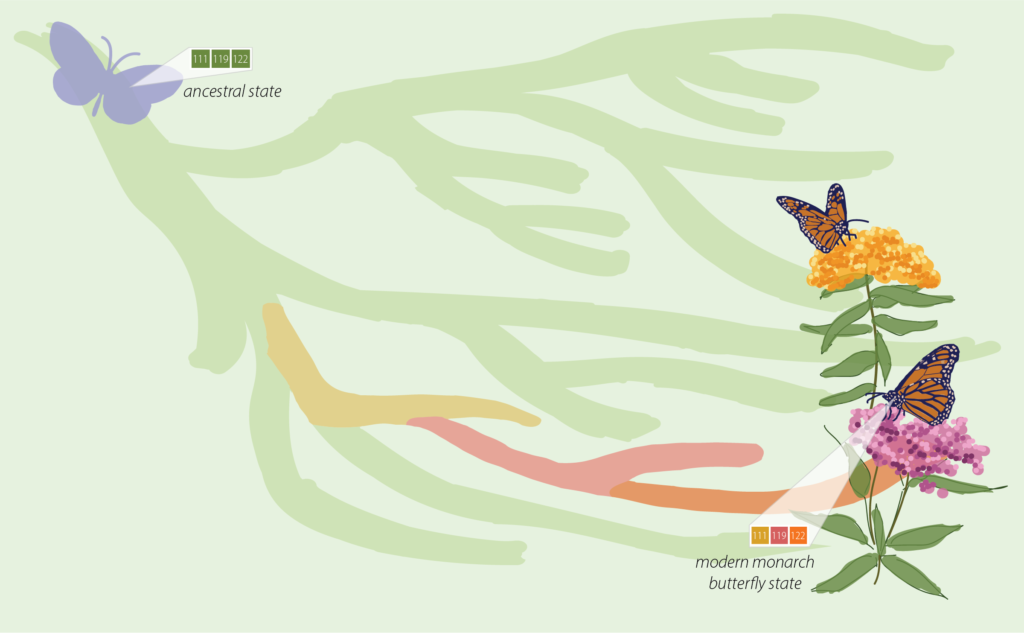 "A hypothesis emerged: mutations at 111 and 122 provided resistance, while a mutation at 119 somehow set the stage for 122, perhaps by counteracting side effects." Image Design: Alison H. Nguyen
"A hypothesis emerged: mutations at 111 and 122 provided resistance, while a mutation at 119 somehow set the stage for 122, perhaps by counteracting side effects." Image Design: Alison H. Nguyen
A hypothesis emerged: mutations at 111 and 122 provided resistance, while a mutation at 119 somehow set the stage for 122, perhaps by counteracting side effects. To test their hypothesis, the team turned to CRISPR, a genome-editing technology for introducing precise mutations into organisms. The plan was to introduce the mutations into fruit flies, which are not resistant to milkweed toxin, and see if they too became resistant.
Lo and behold, the “monarch flies” acquired the same degree of toxin resistance as the monarch butterfly. Perhaps more interestingly, flies that had the mutation at 122 without the one at 119 had severe neuronal defects, demonstrating the necessity of 119 and of mutational order in evolutionary history.
Karageorgi notes that copious data allowed them to generate their hypothesis. “ASR was possible because the sequences of all these insects were available … [Without] such an extensive dataset, we wouldn’t be here.” Similarly, for phylogenetic inference, “more sequence data can generate more robust results,” says Pelaez. “And all the rest of the analyses are dependent on this.”
This article is part of the Spring 2020 issue.