Every year, millions of Americans dutifully line up at their local pharmacy to receive their annual flu vaccine. In contrast to many diseases, the flu gets its own season and countless public health warnings on local television or via targeted advertisements. What makes the flu so special, and why is it the only disease that we have to get vaccinated against every year? After all, the flu is not the deadliest disease, nor the most common, and yet the flu vaccine asks millions of Americans to confront their fear of needles every year. The answers to these questions tell an interesting story of the history of influenza outbreaks and the unique challenges the flu poses for molecular biologists and immunologists attempting to combat it and other diseases.
To understand the challenges of vaccinating against influenza, it’s important to know that the flu isn’t just a particularly severe cold. Many different viruses can cause what we call the common cold. However, the flu is caused by a specific pathogen called influenza virus. When someone gets sick from the flu, their infection has progressed far enough to experience disease symptoms. In other words, you can get infected with influenza without actually getting the flu, which has important consequences for how public health officials design responses to the disease. Mild symptoms of the flu can be similar to the common cold, but more severe infections can have serious health effects and kills tens of thousands of people every year. As a result, public health officials invest huge amounts of time and resources each year to reduce the flu’s impact on society.
At a general level, influenza falls into the category of vaccine-preventable diseases, meaning a person can receive a flu vaccine and then reasonably expect to have a lower risk of getting the flu. An astounding number of lives have been saved since the discovery of vaccines, and pathogens such as measles, whooping cough, and smallpox have been dramatically reduced or completely eradicated from our planet. For many individuals under the age of 40, vaccinations were administered well before we could have had any say in the matter, or really, before we had the ability to say anything at all. The approach of childhood vaccination has worked impressively well. Previously lethal pathogens have virtually vanished from the developed world. All this to say, influenza has a unique place in both infectious diseases and public health that changes how it impacts our lives compared to other vaccine preventable diseases.
Provoking immune responses
Understanding why we receive the vaccine for the flu, but not other diseases, annually, first requires familiarity with how vaccines work. When a person is infected with a disease and then recovers, the immune system kicks into gear to prevent future infections. One way the immune system does this is by developing antibodies, which are small proteins that stick to the surface of the virus or bacterium that caused the disease. If a person gets reinfected with the same pathogen, those antibodies can bind to the pathogen, neutralizing it and halting an infection before it progresses to disease. Critically, these neutralizing antibodies only stick to small, specific parts on a pathogen, called antigens. In most cases, using these molecular calling cards is sufficient for the immune system to prevent infections after it has seen a pathogen once.
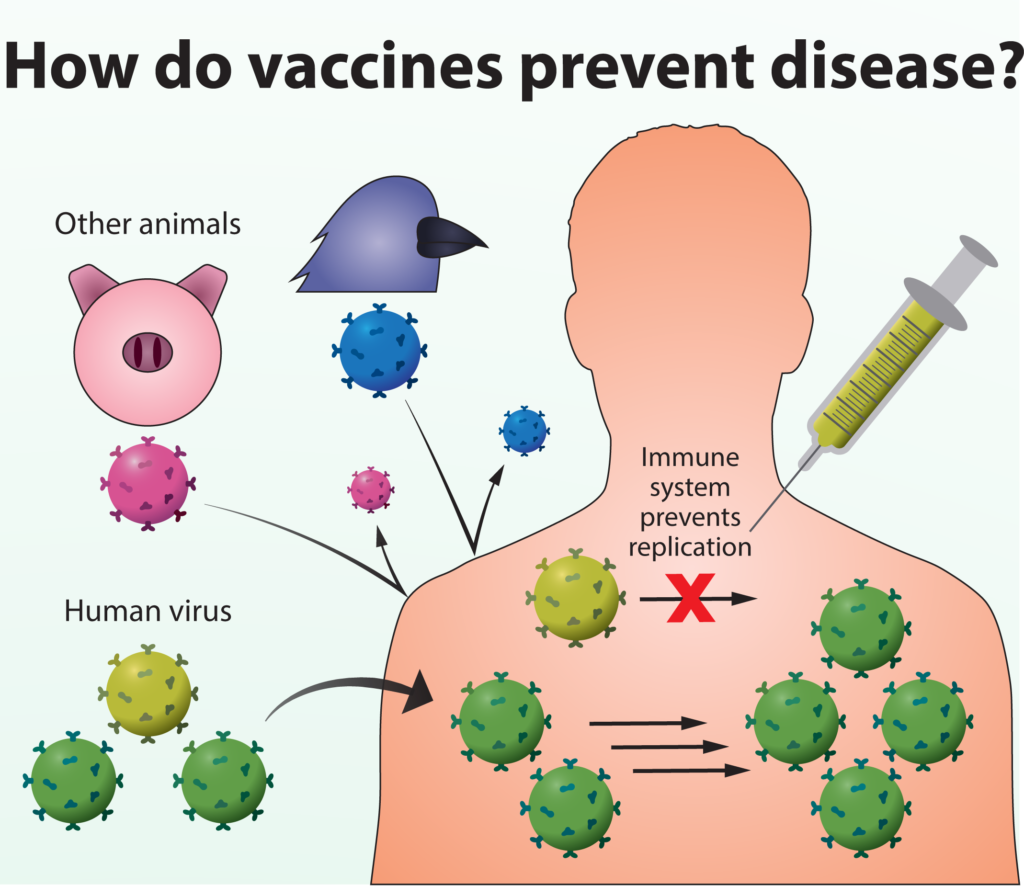 Vaccines do not prevent viruses from entering our bodies, but allow our immune systems to recognize and quickly respond to new infections. The immune system’s ability to quickly recognize a virus as foreign and prevent its replication determines whether or not infection will result in disease symptoms.
Vaccines do not prevent viruses from entering our bodies, but allow our immune systems to recognize and quickly respond to new infections. The immune system’s ability to quickly recognize a virus as foreign and prevent its replication determines whether or not infection will result in disease symptoms.
You can imagine the relationship between the immune system and pathogens as being similar to the way you begin to remember new acquaintances. When you meet someone for the first time, you likely won’t remember their eye color, but might make mental note of particularly distinctive features, like if they had bright pink highlights in their hair. An antigen serves as the metaphorical pink highlights on a pathogen, allowing the immune system to recognize it more easily if it appears in the future.
Vaccines coax our immune system into producing neutralizing antibodies even in the absence of an infection. Simply exposing the immune system to the right subset of antigens allows it to develop antibodies that will protect against a whole pathogen. These antigens can come from a weakened pathogen incapable of causing disease, which is called a live-attenuated vaccine, or by mixing together key pieces of a pathogen, referred to as a subunit vaccine. Both of these methods expose the immune system to antigens and allow it to develop the proper protective antibodies without causing disease. To continue our analogy, it’s as if a friend tells you to keep an eye out for a person with pink hair because they’re planning on stealing your wallet. Even if you have never met this pink-haired person, you have enough identifying information to protect your wallet if they ever show up.
One of the technical challenges of vaccine development is determining exactly what parts of a pathogen to include in a vaccine, and whether they will generate the correct immune response. Arthur Reingold, a professor of epidemiology at UC Berkeley, has spent his career studying vaccine-preventable diseases in the United States and developing countries. He points out, “you can make antibodies [using a vaccine], but they might not protect you clinically if they’re not against the antigens that count.” Thus, vaccine manufacturers must not only ask whether they are producing antibodies against a pathogen, but they also need to know if they’re invoking antibodies specific to the right antigens. The process of choosing the correct targets can be challenging, but the payoff is the opportunity to save millions of lives by stopping infections before they progress to disease.
Tracking a moving antigenic target
These principles of vaccine design are well and good for some pathogens, such as tetanus, diphtheria, and pertussis, which are vaccinated against during the standard course of childhood shots. As a result, these diseases have become historical footnotes, rather than the major public health threat that influenza still poses. One difficulty with influenza, and with many other diseases for which there aren’t vaccines, lies in the specific interactions between a person’s antibodies and the antigens on the pathogen.
Antibodies can only protect against a pathogen if the antigen it initially saw is present during subsequent infections. If that antigen changes, previously developed antibodies are either less effective or completely ineffective at recognizing a pathogen and preventing infection. It’s as if, after your friend notifies you about the serial pickpocket in the neighborhood, you start keeping an eye out for someone with pink hair. You then notice a person with purple hair, politely acknowledge them and internally admire their hair color, before suddenly realizing that your wallet is gone. This failure demonstrates one of the challenges of the immune system’s antigen-antibody strategy: picking one, or even a few, identifying features of a pathogen is only useful if that feature does not change over time.
Influenza virus is particularly good at changing its antigens quickly, which makes vaccine design especially challenging. Professor Jesse Bloom, an associate professor of microbiology at the Fred Hutch Cancer Research Center, studies how genetic variation in influenza virus influences its interactions with the human immune system. He points out that one of the major antigens recognized by the immune system picks up about three changes every year, which he describes as “an extraordinarily high rate, compared to the rate of evolution of most other proteins that we study.” For your immune system, this rate of change means that even if you get infected with the flu and develop a very strong antibody response, those antibodies lose their effectiveness within a few years “simply because the [antigens] that are recognized by the antibodies have changed quite a bit.” The annual influenza vaccine is an attempt to update your immune system as the virus evolves, so it produces antibodies that bind to the altered antigens on the circulating influenza virus.
A major reason that influenza is capable of changing its antigens so quickly is that its genetic code is stored in RNA, rather than DNA. Human cells encode genetic information as double-stranded DNA, which stores information fairly reliably, since the two strands essentially serve as copies of each other. In contrast, the influenza genome is made up of single-stranded RNA. RNA is inherently less stable than DNA, but more importantly, its single strand means that mistakes while copying the flu genome are permanent, and no duplicate exists to correct the mistake. As a result, errors in the genome accumulate rapidly, which in turn changes the antigens produced by influenza virus.
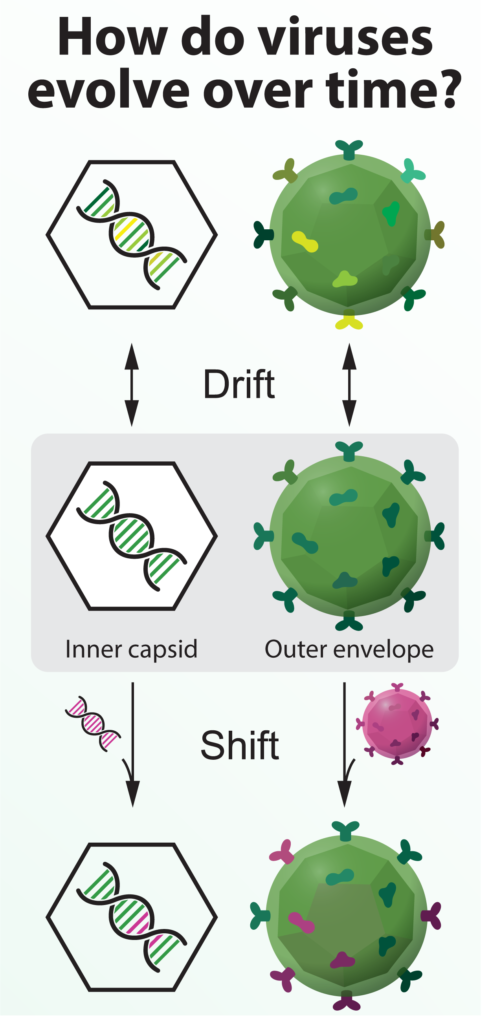 Viruses can change gradually, through the accumulation of genetic errors (drift), or rapidly, by exchanging genetic information with a related virus (shift).
Viruses can change gradually, through the accumulation of genetic errors (drift), or rapidly, by exchanging genetic information with a related virus (shift).
One infected cell can produce hundreds of viruses, each of which has slight genetic differences from other viruses in the cell. This leads to what Bloom calls a “cloud” of very closely related, but genetically unique, viruses. Even if the majority of those viruses cannot grow or infect another cell, a small portion of them will survive with their new mutations. As the virus undergoes cycles of infection and release from hundreds of cells within the body, the cloud of viruses will slowly drift, moving away from the original virus that could be recognized by an antibody. Reingold describes drift as a “gradual accumulation of genetic changes, such that the antibodies or the immunity, whether naturally acquired or vaccine induced, may no longer protect against the strain that’s circulating.” This drift continues as the virus passes from person to person, and your odds of getting the flu again progressively increase as your antibodies become less effective at sticking to the virus.
In order to better understand antigen drift in influenza virus, Bloom’s research group intentionally mutates collections of viruses and examines how they grow in response to different conditions. For example, they can add an antibody to their population of viruses, which prevents antibody-bound viruses from replicating, and then measure what mutations allowed the virus to escape the antibody. In this way, Bloom’s lab can “measure the effect of each of these viral mutations on a property, such as antibody recognition,” and then “build up a sense of which mutations will be of consequence in helping the virus escape from immunity.” A related aspect of the lab’s work uses influenza viruses taken from human patients and measures how the virus changes during a single person’s infection in response to the immune system. By combining these two sets of data, the lab is able to examine a broad range of antigen mutations and compare those with the types of changes that are actually occurring around the globe.
Bloom’s research has revealed a great deal about the processes that drive gradual change in influenza virus. However, influenza also has the capacity to suddenly alter its antigens using a process different from genetic drift. Aside from the viruses that cause disease in humans, there are many other influenza viruses that circulate in animals. Pigs, birds, and horses each have their own influenza viruses. These viruses tend to remain infectious to a single species, with bird viruses infecting birds and pig viruses infecting pigs. However, human viruses can occasionally exchange large chunks of genetic information with bird or pig viruses when a person comes into contact with an infected animal. This exchange changes the antigens on a virus, resulting in an antigenic “shift,” in contrast to the regular antigenic drift. Reingold describes genetic shifts as a “dramatic change in the antigens that are on the surface of the flu virus, as opposed to a gradual evolution.” For humans, genetic shifts mean that most of the population no longer has antibodies that protect against the virus, greatly increasing the chances of infection and disease. Therefore, even the best-designed seasonal vaccine is very unlikely to provide any protection against a flu virus that shifts, since the antigens on the virus change so much.
Research on influenza shift and drift teaches us how the virus changes to avoid the immune system, but transitioning from an annual vaccine to a one-time vaccine will require the immune system to produce antibodies that bind all influenza viruses. Antibodies against an unchanging, or universal, antigen would hypothetically protect a person from every influenza virus year after year, even as other parts of the virus change. In fact, universal antibodies for influenza have been discovered, but scientists haven’t quite worked out how to translate that knowledge to a new vaccine. Bloom explains that researchers have “found antibodies that can recognize essentially all strains of influenza A virus, which is the most common type of flu causing human disease, and these antibodies have actually been isolated from infected humans. They’re very rare, but it shows that they can exist and they can be made.” One of the current challenges is then finding a way to stimulate production of these antibodies in all people—and at a high enough level to prevent an infection with influenza from progressing to disease. Undoubtedly, developing a universal vaccine will be a difficult process, but Reingold comments that, “If somebody figures that out … it’ll be a major public health victory. It may never happen, but we’re certainly working on that. Smart people are working on it.” Until that breakthrough, scientists continue to hunt for a deeper understanding of influenza virus and the way the immune system protects against disease.
Antibody anomalies in dengue virus
Another challenge with the flu is that, even after identifying universal antigens on the pathogen and finding a way to generate potent antibodies, complications may still arise from the human immune system itself. To understand these complications, we turn to a different viral pathogen, dengue virus, which has vexed immunologists for decades even though its antigens rarely change. It is similar to influenza virus in that it is a single-stranded RNA virus, but the two viruses diverge rapidly after that. Influenza spreads directly from person to person, but dengue must be transmitted via mosquito bite to spread to an uninfected person. While influenza has virus types A and B, dengue virus has four different serotypes numbered 1 through 4, that are genetically separate from each other. Dr. Dustin Glasner studied dengue virus with UC Berkeley Professor Eva Harris during his doctoral work and says that while the four serotypes are closely related, “you can think of them like four siblings of the virus.” The disease that people get after infection is initially the same with all four serotypes.
Much like other diseases for which effective vaccines exist, when a person gets infected with one serotype of dengue, the immune system develops antibodies against viral antigens. After this primary infection, a person is theoretically protected for life against disease caused by that dengue serotype. The complexities in designing a vaccine lie with the differences between dengue serotypes. To the immune system and its antibodies, these sibling viruses look somewhat similar, but not exactly the same. When a person gets infected with a different serotype of dengue, the relatedness in antigens means that antibodies against Dengue 1 can bind to antigens on Dengue 2, but they don’t bind tightly enough to block a Dengue 2 infection and are therefore called non-neutralizing antibodies.
Non-neutralizing antibodies have a significant impact on the disease caused by dengue virus—not only because they don’t stop a person from getting sick again, but they also increase the chance of developing an even worse disease. As Glasner describes, these antibodies “won’t clear a secondary dengue infection of the same serotype but will still bind to that virus and can facilitate entry into cells,” making it more difficult for the immune system to respond than if there hadn’t been any antibodies at all. These antibodies that enhance the symptoms of dengue virus present a huge challenge when thinking about vaccine design because the disease caused by a secondary infection is much more severe than a primary infection. In a sense, the immune system has a two-strike policy against dengue virus, where the first infection is rarely lethal, but the second infection has a much higher chance of causing severe disease. As a result, vaccine design against dengue virus needs to avoid swinging and missing, since an imperfect vaccine could put people at much greater risk of dying from a dengue infection.
The lessons from dengue indicate that even if a universal influenza antigen is discovered, there are still nuances in the human immune system that must be understood before it can be safely deployed. Researchers will need to consider how the immune system responds after repeated exposures to different strains of influenza—ideally at both the cellular and population-wide levels. Nonetheless, continued research on influenza virus, dengue virus, and other infectious diseases will help flesh out our understanding of pathogens and the concomitant immune response.
One flu over the chicken’s nest
With all that is known about influenza, combined with researchers’ ability to measure how it changes over time, you might wonder why companies don’t simply update the vaccine as it changes. Outside of the science itself, a number of other factors contribute to making the flu a particularly complex public health problem. To start, about nine months before the flu season begins, a panel of public health officials meet and decide what types of flu to include in the vaccine. They gather as much information about the circulating virus as they can, but they can only make an educated guess as to how the virus will change by the time the next winter arrives. That information gets sent to vaccine manufacturers, who then must grow enormous quantities of influenza virus. For the past seventy years, the strategy for producing massive quantities of virus has been to infect chicken eggs with influenza and then extract the virus from the eggs after they have grown. This procedure adds several wrinkles to the already-challenging process of influenza vaccines.
The first challenge is a question of scale. Reingold explains that “it takes about one egg to make one dose of vaccine. So if you want 100 million doses, you need 100 million eggs.” For each year’s vaccine, companies need to obtain 100 million eggs, inject 100 million eggs with virus, and crack 100 million eggs to get more virus out of them. All of these steps need to proceed under sterile conditions because contamination would have severe consequences once the vaccine is injected into people.
The second problem lies in using the egg as a medium for growing viruses. Human influenza viruses are not very far removed from bird viruses. According to Reingold, “If the influenza virus against which we want to make a vaccine in hen’s eggs kills the eggs, then you cannot propagate it and produce the required amount of virus to make a vaccine.” To get around the problem, scientists might genetically manipulate the virus so it doesn’t kill the eggs. However, influenza’s capacity to rapidly mutate means that the virus might naturally evolve to adapt to conditions in the chicken egg. Such change may lead to a virus that “is not a good antigenic match for the virus that is circulating and causing disease, in which case you can end up making a vaccine that is not very protective against the strain that is circulating,” says Reingold. Theoretically, modern technology should allow us to grow the virus outside of eggs, but a combination of the costs associated with switching over and the regulatory barriers to using new technology has made the transition process slow.
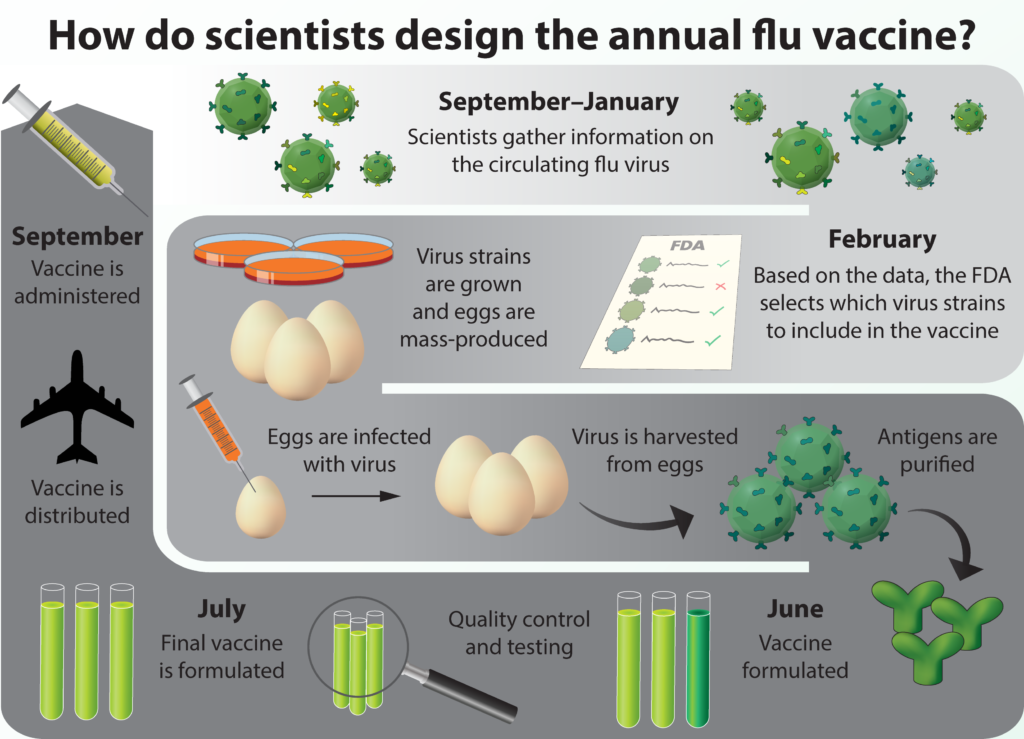 Countless hours are spent designing and producing the flu vaccine each year. While scientists do their best to predict which strains will be present during the flu season, genetic drift and shift cause unpredictable changes, resulting in year-to-year variations in vaccine effectiveness.
Countless hours are spent designing and producing the flu vaccine each year. While scientists do their best to predict which strains will be present during the flu season, genetic drift and shift cause unpredictable changes, resulting in year-to-year variations in vaccine effectiveness.
The cycle carries on
Overall, production of the annual flu vaccine is lengthy and complicated. The entire process, from scientists first picking the viruses to the vaccine ending up in a clinic, takes about nine or ten months and requires participation from multiple different public and private organizations. It demands millions of dollars of investment from governments and pharmaceutical companies, and at the end of the day, its design is simply a very educated guess. On top of that, the flu keeps changing, so as soon as one year’s cycle has finished, the process starts all over again with a new attempt to predict how the virus will drift.
Ultimately, all the work is worth it, as the annual influenza vaccine protects millions of people from disease, but we must still return to the clinic every year to renew our protection with a vaccine that only works some of the time. Reingold acknowledges the frustrating situation, explaining, “most vaccines we use commonly have efficacies in the range of 80-95 percent ... but flu vaccines are typically down in the 40-60 percent range. That’s not very good! And we need to give them every year! So if you say, ‘Is that a good vaccine?’ the answer is ‘No!’” Even if it’s not ideal, the flu vaccine is the best solution available at the moment, and it is far better than nothing at all.
Some sunny day in the future you might be able to visit your doctor for one final shot containing a universal influenza vaccine. With every passing year, we understand a little bit more about the flu and get a little bit closer to a solution. In the meantime, scientists carry on the struggle to vaccinate against influenza, as it ceaselessly changes and outmaneuvers our immune system.
As for what makes the flu so special? Perhaps it’s not its ability to change antigens readily or the breadth of organisms in which it grows, but rather, it’s special just because it’s a problem we haven’t solved yet. Once we do, one hopes that influenza will join smallpox and diphtheria in the footnotes of infectious disease history.
Eric Lee is a graduate student in infectious diseases & immunity.
Design: Matthew Stefely
This article is part of the Spring 2019 issue.