Falling in love with bacteriophage: An interview with Diana Fusco
By QinQin Yu
March 14, 2019
As a young biophysics grad student, I was naturally looking on the internet for resources to help give me context for what it’s like to work in this interdisciplinary field and what quirks of the field I should expect. I was surprised to find very few resources, and even fewer personal accounts, from biophysicists themselves (see the bottom of this article for some examples). I decided to interview some biophysicists in my own lab to get some insight.

This first interview is with Dr. Diana Fusco, who has just started a research lab as a Lecturer of Physics at Cambridge University in England (equivalent to an Assistant Professor position in the U.S.), after recently wrapping up a postdoc in Professor Oskar Hallatschek’s lab at UC Berkeley. Below, we hear about her current work on evolution of viruses, her nontraditional path into biophysics, and her advice for those new to the field. Some text from the original interview transcript has been edited for clarity. The interview was conducted on June 13, 2018.
QinQin: Could you tell us a bit about what you’re currently working
on?
Diana: The thing
that I’m most passionate about and also envision the future of my lab’s
research [focusing] on are bacteriophages, particularly evolution of
bacteriophages. What’s a bacteriophage?—because not everybody may know. We
often think of viruses like influenza or Zika virus, which affect humans and
animals, but the bacterial world has its own viruses. Bacteriophages are the
flu of E. coli and other kinds of
bacteria. What I study is how these viruses evolve to learn how to infect
bacteria, in particular E. coli,
especially when both the viral and the bacterial population are distributed in
space. So this is in contrast to when you mix all of them in a test tube and both
the viruses and the bacteria have access to the same environment. Whereas, when you have them [confined] in
space, each of them is in a heterogeneous environment and will feel different
conditions. That’s generally what I study and what I’m going to study in the
future.
QinQin: Why do
you think this question is important for the real world?
Diana: That’s a good question. I feel like what drives me to [study] these questions is often not why they’re important in the real world. It’s just because they’re fascinating [to me] for some reason. But in the case of bacteriophage, it is one of the cases where I think it’s really important in the real world for multiple reasons. Bacteriophage is the most common entity on the planet. If you think …[of] the whole spectrum of organisms on the planet …, bacteria are [one of] the most abundant. For each [species of] bacteria, you have ten different species of bacteriophage, so they’re really big in that sense.
In the wild, when you go to just pick a glass of water in
the lake, or [collect] a piece of soil in the ground, you’ll find that actually
bacteria and phage coexist and … bacteriophage is responsible for most of
what’s called horizontal gene transfer among bacteria. Most bacteria reproduce
asexually, so they can’t mix their genetic information like people, [where] you
have mother and father together, they make a child, and the child will have
genetic information from both of the parents. For bacteria that’s not the case.
You are identical to your mother. From an evolutionary standpoint, this is a
bit disadvantageous, because it means that you can get stuck with bad mutations
and they make you less fit. So it’s kind of nice to actually have a way to exchange
genetic material between individuals. Bacteriophages, like viruses, allow
transfer of genetic material between individuals. A way to think about them,
almost a physics way of thinking about them, is as messengers of information in
the bacterial world. And they’re really responsible for 90 percent of the horizontal
gene transfer in the wild.
Even in our gut, the microbiome now is really important. In
that case, phage plays an important role for similar reasons. One of the
current problems in medicine is resistance to antibiotics in bacteria, and
phage therapy was proposed some decades ago [as a potential solution to this
problem]. It’s kind of gone out of fashion, actually, but is having a comeback
as a way to support antibiotics when you get an infection and you need to treat
it. Sometimes the antibiotics may not work because the bacteria may be
resistant. Using phage to kill the bacteria is an alternative to antibiotics,
or something that can support antibiotics. Some people are really looking into
these options.
So this is why phage is really important. Of course, a lot
is known in the ideal conditions of how these viruses evolve, but if you want
to really look at them in real conditions where space, like who interacts with
whom, becomes really important, you need to think about what those constraints
are going to do to the evolution of the viruses and the bacteria, because they
both evolve very fast.
QinQin: What
motivates you study these questions?
Diana: Hard to
say. I think it’s almost like when you fall in love with a person. I can give
you my experience. Though I liked biophysics, I cannot say that before my
encounter with bacteriophage I was really passionate about something. There
were all these questions [which were often] my advisors’ questions. I thought
they were interesting. That’s why I did research on them. I thought how to
expand them, mostly how to get papers out of them, because that’s what you
always think about. But I was never really thinking, “Oh my god, why does this
happen?” It was never [that I] was really curious about these things, it was
just like that’s my job and that’s what I study, and that’s it.
Instead, my encounter with bacteriophage was purely
fortuitous. We had a visitor coming into the lab who worked with bacteriophage.
I just started looking at what he was doing and playing for a bit and then it
became like almost an obsession, because it was like, “Oh my god, but they are
so small.” [The] genetic information in bacteria is like 1,000 genes … and
these phages can be a [hundredth] of what the bacteria are [in genome size].
The information is really … compact there. And still they are able to survive
in many, many conditions, adapt in all kinds of conditions, and act on hosts
that they get exposed to. It became like, how is this tiny, tiny thing able to
do all these things? For me it was crazy. Within our current world … your
operating system by itself takes up terabytes—well I’m exaggerating, you know
what I mean—and this tiny thing has been on this planet forever, and they can
do all this. How can they do that? One question fed the other …. They have to
pack all this information in the genome. They have it packed where the specific
order of how these things are put together matters. If you switch the order,
then things change. How did they come up with this? And it starts hitting all
kinds of questions, and it becomes almost like a mystery. You have a specific
question maybe you want to ask, and you look for clues and for ways to interrogate
your system to find what the answer to the question is. What drew me exactly? I
think it was that I was fascinated by how such a tiny, tiny system can do so
much, and then it just exploded in billion questions.
QinQin: You
already talked a bit about how you arrived at studied bacteriophages. Could you
talk a bit more in detail about how you got involved in biophysics in the first
place?
Diana: The first
question is, what’s biophysics? I think that’s really a big question. I don’t
have the definition to this. My way of thinking about it is that it’s a way of
thinking about biological questions by using a more physics way of asking
questions somehow, where you really look more for … universal features that can
encompass many systems and can explain a lot of features, almost like a coarse-grained
view of what’s happening out there in nature. That’s one definition. I think
there are other definitions. That’s kind of what drives me there.
For me …, maybe because I was trained as a physicist, I do
think that there are some general rules out there that govern also biology in
all its craziness and exceptions and everything. I think if you zoom out you do
find these general patterns. These general patterns, and looking for general
patterns, was what drew me to physics to start with.
As an undergrad, I actually hated biology. I thought it was
like this list of names and things, and my memory is awful. Things that I would
never remember. And then I came upon biology, transcription factors, and networks.
During my master’s, there was a professor that I wanted to do my dissertation
with and he proposed this project to me that was really very much like a
computational mathematical kind of project. It was like finding modules in a
network or something. The only biological thing was that the network we were
studying was a biological network. Once I restarted reading more papers I
realized, “Oh, this biology thing was not so bad.” I felt my fear was really
that I was thinking about biology more like how it was in the old days, before
molecular biology became a thing. It was maybe just a list of species or
features of different animals and things like that. Instead now, it became
something that I feel you can actually question and things you can measure that
you can answer in a very quantitative way. That’s where I thought, “Well, maybe
it’s not so bad.”
After that I said, “Well, I’ll try to do a PhD in
computational biology. Why not? I’ll see what’s out there in this computational
biology thing.” I remember at the time that I applied, I really thought that
structural biology and computational biology were the same thing. I was using
them interchangeably. And then I found out, oh no, structural biology is all
about how the molecular structure of how these things are; instead,
computational biology is also that and many, many other things. That was my
understanding of how these things were. It was very, very coarse and unclear.
Somehow I managed to start a PhD in computational biology.
In the end it was really good because it exposed me to many
different biological questions, but all asked more in a quantitative way. I was
studying genomics, so how changes in the genomes, like conformational changes,
affect the organism. That was more systems biology. That was looking at all
these things put together. There were many, many of these questions and I
fluctuated a lot. Initially, I got more into evolution, I think, because I was
always kind of interested in evolution. But then I had kind of a bad experience
with my first PhD advisor, so I decided “Well ok, maybe this is more
biological.” The step was too big and I shouldn’t have done that. After all, I
was a physicist, so maybe the biological world was not for me.
So I stepped back and went more into soft matter, which is actually
physics, I would say, but with a lot of applications in biology. In my case,
the application was protein self-assembly. That was what I was studying for my
PhD. I really felt, at that point, that I was doing physics, just applied to
biological systems. I didn’t care that it was applied to biological systems; it
could have been applied to anything. What I cared about was more the tools.
After that, at the end of the PhD, I felt, “OK, this is
enough. I actually want to try this biology thing again and see whether I can
give it a second shot,” and that’s when I joined the Hallatschek lab. Especially,
I think for me, [in my PhD lab,] one of the big drives was that I was doing
only computational and theoretical stuff, and I got very frustrated with my
computational work. We had all of these theories which no [experimental] lab
wanted to prove because no one wanted to do the experiments for us, and I
thought, “I want to learn how to do the experiments.”
So I spent a lot of time reading around for what kind of
experiments [I wanted] to do, because there’s a lot of stuff out there. Given
that my PhD expertise was more on the molecular scale, I was trying more to get
into a molecular biology kind of lab until I found out that for a person with a
purely computational PhD, when you look for a postdoc, the PI may not be so
keen to have a person with so little experience in doing experiments at the
postdoc level. They will invest the time of course for a PhD student, but for a
postdoc, they want to see that you can hit the ground running pretty much. I
kept looking and I thought I would also be interested in the larger length
scale, like organismal level. That’s how I got more into microbiology and I
came across Oskar [Hallatschek] by chance. I was just googling people and it
came up. Somehow, Oskar was very responsive to having a person like [me] because
he kind of followed a similar path himself [from theory to experiment]. He was
willing to have a person that had no idea of how to even hold a pipette. I
think that really was the game changer.
Once I actually started doing experiments myself, that
really triggered my curiosity as well. You see the things happening. I feel
like now I’m almost more and more into the experimental side and kind of left
the theory. I still do theory but it’s more like, “I have to do it because I
still have to understand this thing. I still want to understand what’s
happening.” I really use it as a tool, but the driving question is
experimental.
QinQin: I can
imagine for many students out there, it’s a dream to be able to find something
that motivates them and gets them excited. I know you said that you’re not
really sure how it happened, but do you have advice for students that really
want to find these questions?
Diana: I think
first of all [I would tell them] to not to be afraid of exploring many
different things. There are pros and cons in being too spread in interest,
because when you do your PhD, you become super specialized and you become the
expert in the world for that super specific thing. I would say it’s a great
thing to explore things outside your specific area, because you never know where
you’ll find the thing that excites you.
My advice is … even if I think that I studied quantum
physics for 10 years, and I’m going to do astrophysics now, well, it’s fine.
It’s fine to jump. One should not be afraid to do it. It may mean extra work at
the beginning because you have to catch up, and you may feel a bit lost at the
beginning because you feel that everybody else is understanding what the heck
is going on here because they’ve already studied this for several years and
you’ve just started. But you’ll get to it. Just don’t be afraid of that. That’s
my suggestion. You’ll never know [when] you’ll find an interesting thing.
Find a good lab and a good mentor. That will make your
experience so much better. It nourishes you, so you feel part of a community,
and I think that’s really important. Sometimes, I feel it’s more important than
what your research actually is on, … that you want an advisor that is
supportive of whatever decision you decide to make afterwards. I stand by what
I said, just be happy. If at some point you feel miserable, you’re not meant to
feel miserable, so it means that there’s something wrong. There’s nothing wrong
with changing and doing something that doesn’t make you feel miserable but
makes you feel happy.
Check out these other blogs about biophysics:
Raj Lab for Systems Biology Blog
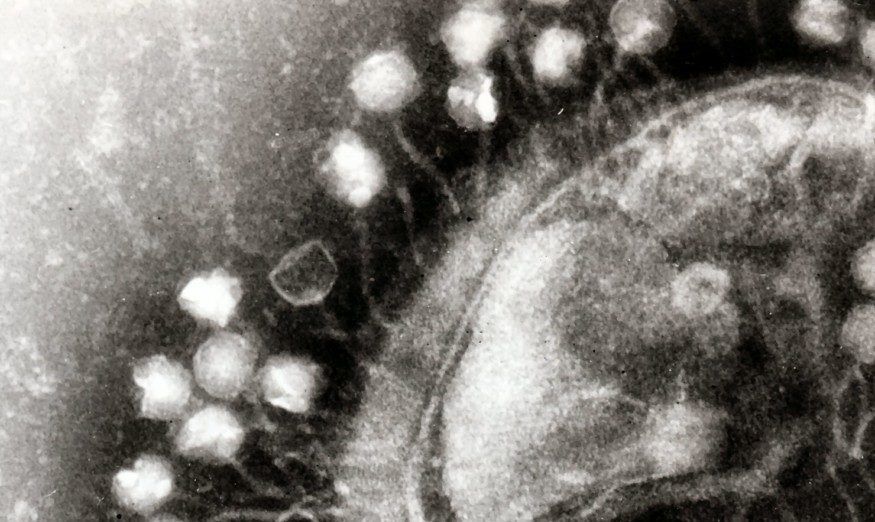
Featured image: Transmission electron microscopy image of multiple bacteriophages attached to a bacterial cell wall. Each bacteriophage is about 100 nanometers. The image has been cropped from the original. Source: Dr. Graham Beards