Extremophiles are organisms that thrive in places where most life would perish, including the coldest, driest, and most acidic or basic environments on our planet. Researchers use an arsenal of experimental techniques to illuminate the strategies by which life persists in these harsh conditions. The field focuses on a central conundrum: how do extremophiles circumvent the laws of chemistry and physics to survive?
The drought-proof organism
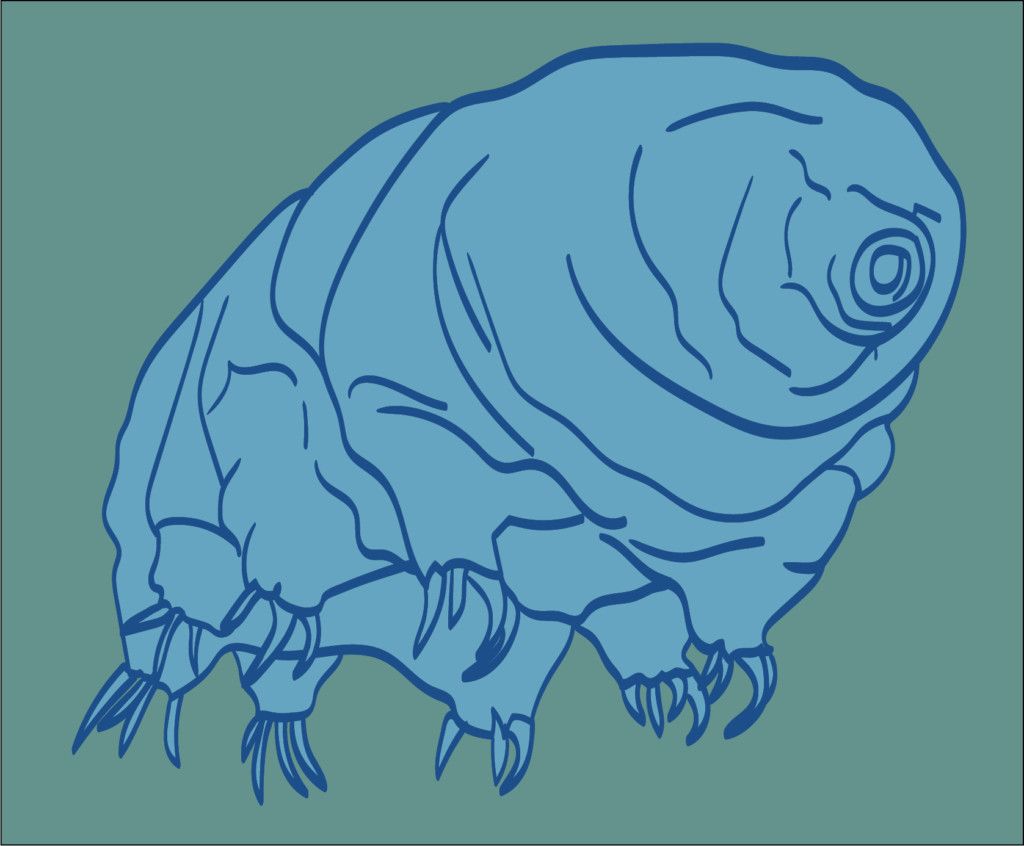
Microscopic, plump, and lumbering, tardigrades are not obviously impressive animals. But don’t be fooled—they are incredibly tough. Tardigrades tolerate deep freezing, total dehydration, bombardment with DNA-scrambling radiation, and the vacuum of space.
Recently, biologists at UC Berkeley and UNC Chapel Hill described a particular flavor of protein that is vital for tardigrades’ ability to survive dehydration (also known as desiccation). Tardigrades possess a unique protein that is unusually flexible and fails to conform to a particular shape. Without these proteins, tardigrades lose their famous ability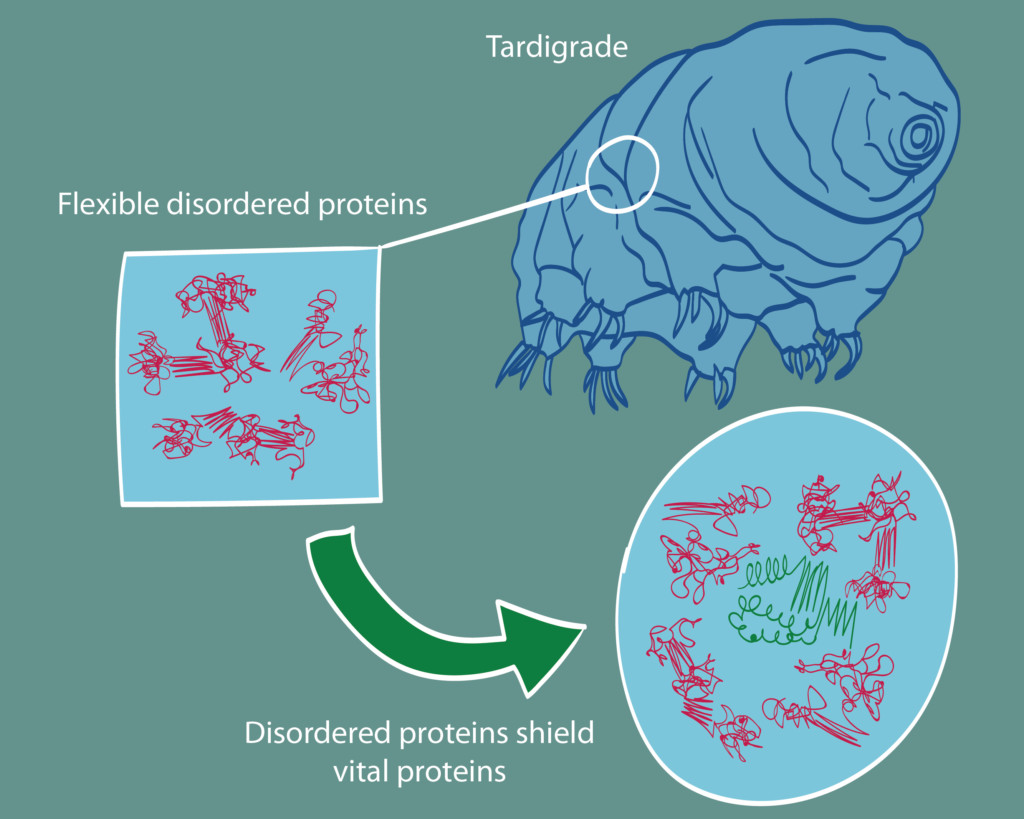 to survive desiccation.
to survive desiccation.
Exactly how these ultra-flexible proteins allow organisms to live without water is still a mystery. One theory is that the disordered proteins (magenta) form a flexible shield around other vital proteins (green), preserving them during desiccation and then releasing them when the tardigrade is re-exposed to water. Alternatively, the disordered proteins may be fulfilling the chemical role of water by stabilizing hydrogen bonds, the tape that holds important proteins in the right shapes.
Researchers are now interested in using these proteins to preserve sensitive drugs or engineer drought-resistant crops. Aside from Earth-bound applications, the mysterious mechanism tardigrades use to survive extremes has stoked the imaginations of astrobiologists (see “Life on the Edge”, page 12), hinting at the potential for life on planets with extreme variations in water.
A heat-proof bacteria
Characterizing the most extreme life on Earth is often more than curiosity-driven. Occasionally, these exploratory investigations uncover molecular tools that prove massively useful to scientists.
In 1969, while on a hunt to collect and study inhabitants of the scalding hot springs of Yellowstone National Park, biologists isolated a particularly heat-tolerant species of bacteria and named the species Thermus aquaticus. These bacteria are skinny, rod-shaped organisms that thrive in temperatures as high as 80°C (175°F).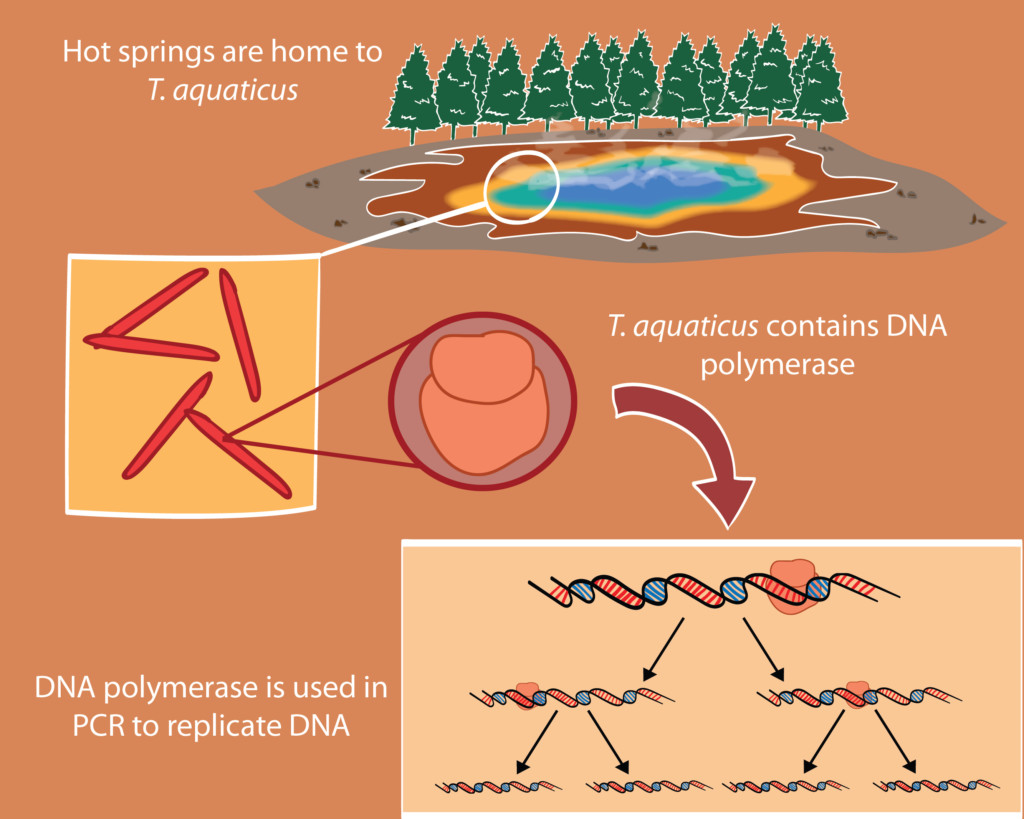
Living at high temperatures puts strain on vital biological processes. Genome replication must proceed with minimal error, or fatal mutations might be introduced into the DNA sequence of the organism. The process relies on a protein called DNA polymerase, which usually unravels at high temperatures. T. aquaticus has an unusually heat-stable DNA polymerase, allowing it to withstand the heat and replicate its DNA without trouble.
Biochemist Kary B. Mullis, working at Cetus Corporation, harnessed the temperature-stable properties of the T. aquaticus polymerase to build what can be thought of as a chemical Xerox machine for DNA, using cycles of heat and cold to amplify specific DNA sequences. The procedure is termed the polymerase chain reaction (PCR), and the impact it’s had on molecular biology is hard to overstate. PCR is routinely used in genome sequencing, to identify rare cell types, and to track amounts of gene expression. The discovery earned Mullis the Nobel Prize in chemistry in 1993.
Maya Emmons-Bell is a graduate student in molecular and cell biology.
Image credit: Jo Downes Bairzin
This article is part of the Fall 2017 issue.



