DNA in bacteria is constantly changing shape—it recombines, replicates, and compresses into organized structures. This remarkable plasticity would be impossible without helpers known as nucleoid associated proteins. Recently, Dr. Michal Hammel, a scientist at Lawrence Berkeley National Lab, headed a study aiming to understand how DNA interacts with these nucleoid associated proteins to create tightly compressed DNA structures. The study used a technique that reveals structure on the scale of a single atom, known as X-ray crystallography. By crystallizing short DNA fragments with the nucleoid associated proteins, Hammel and his research team discovered regions of the proteins that distinctly bind to DNA. These packaging proteins formed more complex networks and structures with the DNA than previously thought.
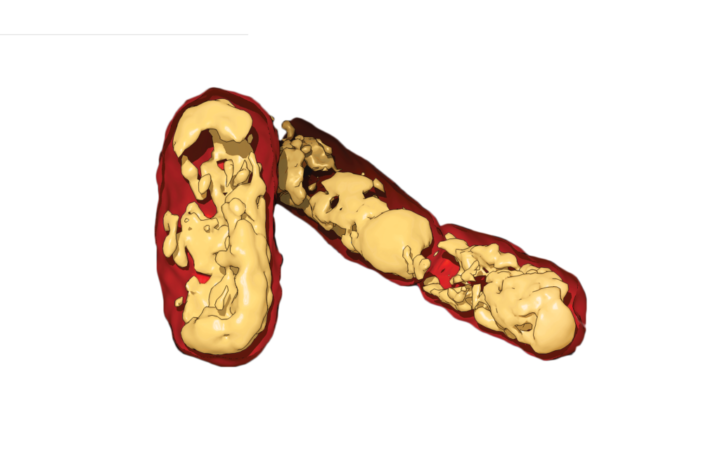 3D reconstruction of E. coli from soft X-ray
3D reconstruction of E. coli from soft X-ray
While X-ray crystallography is a powerful technique, artificial laboratory-grown crystals sometimes do not realistically reflect biology. The team sought to confirm their findings in living bacteria using an additional imaging technique called soft X-ray tomography, which bombards bacterial cells with X-rays to produce flat 2D images. The 2D images are then combined computationally to create a 3D picture of the whole, living bacteria. When packaging proteins lack the DNA-binding regions, the team found that the previously observed DNA networks are missing. In fact, the bacterial DNA seems to collapse into disarray, and the cell no longer grows properly. The team anticipates that these new findings may eventually lead to the development of next-generation antibiotic drugs that target this packaging system.
Soumya G. Remesh is a postdoctoral fellow in molecular biophysics & integegrated biology
Design credit: Kurtresha Worden
This article is part of the Fall 2017 issue.