The past year saw a high profile failure in the emerging field of point-of-care diagnostics. Here’s a science-focused take on why.
In October of 2015, biotechnology enthusiasts and investors across Silicon Valley reacted with varying degrees of shock and disappointment as the Wall Street Journal reported that promising diagnostics startup Theranos was hiding serious flaws in its core technology. Former employees of the nine billion dollar-valued company, headed by Jobsian wunderkind Elizabeth Holmes, alleged that the vast majority of the tests performed by Theranos were conducted using traditional blood testing equipment and that the company was hiding negative results. At the same time, investigations of the company by the Food and Drug Administration (FDA) and the Centers for Medicare and Medicaid Services (CMS) were turning up a lack of regulatory compliance and poor quality control.
This was the beginning of a hard year for Theranos, as further tough press about unfulfilled promises and a poor working environment accompanied the dissolution of their partnership with Walgreens in June. The coup de grace came in early July, when the CMS, based on the results of their 2015 investigation, issued a list of harsh sanctions against the company, which cut Theranos off from critical Medicare reimbursements and banned Theranos and Holmes herself from operating clinical laboratories for two years. Enthusiasm for the company among investors, scientists, and the press evaporated away; in a move emblematic of the changing mood, Forbes reduced their estimation of Holmes’s net worth, mostly composed of the value of Theranos, from $4.5 billion to “nothing.”
Why did the story of Theranos’s miracle finger prick, which seemed from the outside like it had all the makings of a breakthrough saga reminiscent of the early days of computing, turn into a cautionary tale for biotech investors? There are no shortage of articles blaming the culture of Silicon Valley and the secretive environment Holmes implemented at her company. However, there is another part of the explanation that has nothing to do with business practices, corporate culture, or even bioethics: their device didn’t work because it’s a beast of a cross-disciplinary engineering problem. Designing products like Theranos’s proposed device is not impossible, but it is very, very complicated. The rest of this article will attempt to summarize a few of the reasons why.
On the shoulders of (micro-)giants
Holmes’s dream is built on an older idea. In 1990, an analytical chemist named Andreas Manz proposed the idea of a “miniaturized total chemical analysis system” or “micro-TAS,” an integrated fluidic-electronic system that would combine sample preparation and highly sensitive chemical detection in a single device. Manz and other thinkers, inspired by the rapid development of miniaturized electronics, realized that features inherent to the physics of chemical mixtures meant that huge gains in detection speed and sensitivity could be realized by shrinking the fluid-carrying channels down to very small diameters. These tiny “microchannels” could be hooked up in parallel to quickly analyze large volumes of chemicals. Alternatively, small numbers of channels could be used, allowing for the feature of microscale diagnostic systems that appeals most viscerally to would-be patients: the requirement for only a small volume of fluid (such as a single drop of blood) to yield results.
As breakthroughs in microfabrication were driven by demand for faster computer chips, academic enthusiasm for the micro-TAS idea grew. Research labs in physics, chemistry, biology, and engineering developed an increasing variety of miniaturized fluid handling chips, now commonly referred to by the catchier handles “lab-on-a-chip” or “Bio-MEMS.” (“MEMS” stands for “micro-electromechanical systems,” a class of electronic devices that includes the tiny gyroscopes and resonators in your phone.) Microfluidic channels as small as a few thousandths of millimeters (micrometers) in diameter, many times smaller than a human hair, were etched in silicon and glass, leveraging the semiconductor engineer’s familiarity with these materials. More recently, the field has switched to using low-cost and easily handled polymers; glass and silicon processing relies heavily on nasty chemicals such as hydrofluoric acid, which you may remember from its evocative, if somewhat inaccurate, cameo as a body disposal method in Breaking Bad.
Particularly popular among current research labs for its ease of use is silicone rubber, which can simply be poured over an intricately detailed mold and peeled off after it hardens, yielding a rubbery slab with a channel network imprinted in one side, a process known as “soft lithography.” This appealingly simple and robust process has made it possible for scientists without advanced nanoscience knowledge or cutting-edge facilities to participate in lab-on-a-chip research. However, the success of the field outside academia is limited, in stark contrast with the exponential growth in the microelectronics industry. To understand why, we can turn in part to the science, specifically to the fields of fluid dynamics and microfabrication. In other words, we need to talk about the plumbing.
The red stuff
If human biology had, for some reason, been designed with the objective of making it easy to create portable blood tests, the engineer in charge of blood would probably have been fired. According to Kathryn Fink, a recent PhD graduate from UC Berkeley and an expert on the dynamics of blood flow in microchannels, the wonderful properties of blood as a circulatory fluid are a double-edged sword for microfluidics engineers:
“Many of the properties of blood that benefit us physiologically present unique challenges in microfluidic devices. Specifically, red blood cells, which represent about half the volume of our blood, are great for oxygenating our tissues but are tricky in microchannels. Our red blood cells have a strong tendency to cluster together at low flow rates, which is the perfect recipe for clogging small device channels. Circumventing this problem by increasing flow rates leads to some interesting fluid dynamics as a result of the cell deformability: the viscosity varies with volume fraction of red blood cells, how deformable the cells are, and the size of the microfluidic channel.”
In short, the ubiquitous red blood cells tend to muck up our plans, if not by sticking together and to channel walls and creating an impenetrable clog, then by throwing off the assumptions that make it possible to mathematically model fluid flows. To understand how a fluid behaves, we use equations that predict measurable properties of the flow, like the pressure and the flow rate, from the material properties of the fluid, like its density and viscosity. Many common engineering fluids like water and air are easily modeled as Newtonian fluids, meaning that their viscosity is independent of how fast the flow is moving. Even fluids that have a more complicated relationship between viscosity and flow rate can often be described very accurately as shear-thickening or shear-thinning fluids, meaning their viscosity increases or decreases with the flow rate, respectively.
Blood defies such pigeonholing. Its viscosity can both increase and decrease with flow rate, and also unpredictably with temperature, channel size, concentration of blood cells, and so on. In a full-scale lab, this problem is usually solved by centrifuging the cells out of blood before running tests, leaving you with the cell-free fluid (called blood plasma). For a self-contained “total analysis system,” however, the filtration needs to happen on chip – another difficult step in an already complicated device.
Mixing it up
After the blood problem is dealt with in the first stage of the device (usually called “sample preparation,”) the next task is to carry out whatever detection reaction allows you to sense something interesting about the sample, and this isn’t exactly the easy part. Even if the chip carries nothing more complicated than water, fluids at micron-sized length scales tend to behave in ways that we macro-sized humans don’t expect. Of significant consequences for anyone trying to, for instance, combine a patient blood sample with a test drug, fluids in tiny channels don’t like to mix. Mixing two fluids – like milk and coffee, to use an example that most readers of this blog are likely familiar with – is most easily accomplished by stirring or shaking the fluids, creating rapidly moving currents: turbulence. These chaotically moving streams quickly combine coffee and creamer to create a homogeneous, light-brown, less acidic-tasting beverage.
Turbulence is a result of inertia, which you may know as Newton’s First Law and the reason why you need to have brakes on your car. Large amounts of fluid moving fast, like air over an airplane wing or water through whitewater rapids, have quite a bit of inertia and therefore produce turbulence. However, flows in small channels have negligible inertia – it is cancelled out very quickly by the drag forces from the walls of the channel, which are only microns away, so turbulence doesn’t happen in micro-flows. This means that mixing at the microscale relies entirely on diffusion, the slow spreading out of molecules by thermal motion. To see for yourself how slow this process is compared to turbulent mixing, carefully add a single drop of creamer to your cup of coffee and don’t disturb it. You can see the drop slowly spreading from gravity and diffusion.
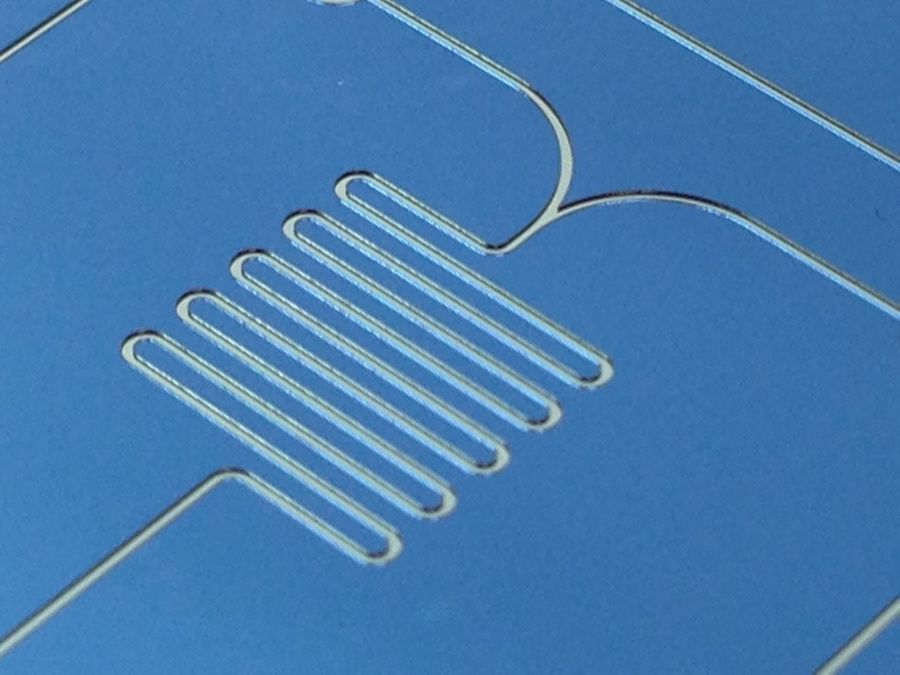
Researchers have worked on a variety of ways to speed up this slow process, but the most common and direct method is just to add time. Those long, serpentine patterns in many microfluidic devices, like the one pictured above, aren’t just there to add baroque decoration to labs-on-chips (though they do make for appealing journal cover photos) – they give streams of molecules time to slowly mix.
Material concerns
After the design difficulties with sample preparation, mixing, and a variety of other complications from which this article will spare the reader, the device needs to make it out of the lab and into production. While academic labs are excellent at dealing with complex cellular interactions and weird fluid dynamics, this is the stage at which the vast majority of microfluidic devices fail to make the cut. A chip designed to process potentially hazardous patient samples in the field needs to be light, cheap, and disposable. The materials used to prototype Bio-MEMS devices in academic labs, glass and silicone rubber, aren’t very well suited to this problem: they’re expensive and time consuming to use.
Luckily, there exists a material that is cheap, easy to mass-produce, and capable of patterning with tiny features, and to find it, we only need to look as far as the humble Lego brick. Though the bricks are made of inexpensive plastic, their shape must be incredibly precise in order to fit together, and accordingly the technology used to make them, called injection molding, is accurate down to two micrometers, more than small enough to mass-produce microfluidic channels. Injection molding involves, not surprisingly, injecting molten plastic into a mold in the shape of the part and then letting it cool, and is unparalleled for producing hundreds of thousands or millions of parts out of plastic for cents per part.
Unfortunately, it doesn’t make much sense to shell out hundreds of thousands of dollars for a precision mold and an injection molding system to make a few prototypes for lab tests, so while injection molding is a manufacturer’s dream, it’s not in most researchers’ toolboxes. Instead, researchers use rapid prototyping methods like the aforementioned soft lithography, which are incompatible with the plastics injection molding requires. This material mismatch between prototype and product can cause quite a few problems as cells, chemical reagents, and fluids can behave unpredictably when the switch is made. New advances in rapid prototyping methods for plastic, such as 3D printing, hot embossing, and micromilling, are steps toward solving this problem, but it remains a huge obstacle in getting the chip out of the lab and into clinics.
The way forward
Though the problems faced by would-be portable diagnostics innovators are complex, progress is still being made toward the dream of ubiquitous point-of-care devices. A variety of startup companies at various stages of development are currently seeking to use microfluidics technology for point-of-care biosensing, many of which use disposable plastic chips. A related technology, called “paper microfluidics,” takes advantage of capillary action to drive flow through paper strips with patterns of biomolecules printed on them, doing away with many of the problems faced by more complex devices. Paper microfluidics could be considered the oldest form of Bio-MEMS technology; home pregnancy tests, a mainstay of drug store shelves for decades, use the same principle.
A particular focus of recent point-of-care diagnostics research has been on emerging global health problems such as Zika virus, against which the ability to deploy inexpensive field diagnostics is obviously indispensable. The cost and portability requirements for such applications are extreme, and in cases where conventional lab tests are not easily miniaturized, Bio-MEMS researchers are working with biochemists to develop alternatives: for instance, isothermal DNA amplification techniques as an alternative to PCR.
Even Theranos itself, almost a year after the first disastrous Wall Street Journal story opened the floodgates, is still moving forward with a new device, which they call the miniLab, in a strictly research and development context (steering clear, of course, of performing clinical diagnostic tests on humans). While the dream of a suite of pinprick blood tests for every major disease is almost certainly out of their grasp, it lives on elsewhere, and advances in micromanufacturing, physics, and biochemistry are leading us there one step at a time.





