It’s lunchtime in the year 2050 and you’re grocery shopping for a snack. Sandwich or salad? Sesame bagel or that gourmet cheeseburger you’ve been craving? To aid your choice, you decide to go with your gut….literally. Within minutes, you’ve consulted an app on your phone which lists your current intestinal microbe composition. Looks like you should go for that salad; your ratio of Firmicutes bacteria vs. Bacteroidetes bacteria is high and elevated Firmicutes levels are correlated with obesity. With complete confidence, you order your salad with a probiotic Bacteroidetes booster, knowing that your choice will temporarily restore your flora to a more lean balance. While this scenario is currently science fiction, recent research at UC Berkeley indicates that a world of personalized “guts guidance” may not be far off.
The human gut or gastrointestinal tract contains over 100 trillion microbes, the largest concentration of microorganisms in the human body and ten times greater than the total number of human cells (1,2). Originally thought to be harmful to humans, we now know that these microbial communities or “gut microbiome” coexist peacefully with us in a symbiotic partnership that coevolved over thousands of years. Indigenous microbes benefit us in many ways including the release of critical nutrients from our food, providing a physical barrier against harmful invasive species, guiding our immunity and aiding in the development of the nervous system (3,4).
Genetic sequencing of DNA extracted from human fecal samples and intestinal tissue indicate that bacteria comprise 99% of this community yet only 20-30% of people with healthy gut microbiome encoded genes are shared between individuals (1,3). This large interpersonal variance in microbe abundance and composition has lead researchers to propose the existence of a “gut fingerprint” or unique microbial signature (5). Identifying and tracking such a signature over time could provide a window into human health and disease and has broad implications for the development of personalized medicine and lifestyle choices.
Research at UC Berkeley has highlighted the influence of age and geography on gut microbiome development (8-12). The dominant bacterial groups in the gut are Bacteroidetes and Firmicutes (1,2,4).
[caption id="attachment\\_12833" align="aligncenter" width="600"][](http://berkeleysciencereview.com/wp-content/uploads/2015/06/gut2.png) Bacterial composition of the adult gut. Bacteroidetes bacteria and Firmicutes bacteria (in bold) are the most dominant bacterial groups. Modified from Cho I, Blaser MJ. (2012).The human microbiome: at the interface of health and disease. Nat Rev Gen. 13(4), 260-270.[/caption]
Elevated Firmicutes and decreased Bacteroidetes levels are correlated with obesity in both mice and humans (6,7). Taichi Suzuki, a graduate student in the Department of Integrative Biology, questioned whether changes in gut composition could mimic the geographic association of large body mass with increased latitude. In conjunction with Dr. Worobey at the University of Arizona, Suzuki analyzed gut microbiome data from over 1,000 individuals in Europe, North America, South America, and Asia (8). The results were stunning: obesity associated Firmicutes positively correlated with latitude while Bacteroidetes levels negatively correlated with latitude regardless of age, sex, race, or bacterial detection methods.
[caption id="attachment\\_12835" align="aligncenter" width="600"][](http://berkeleysciencereview.com/wp-content/uploads/2015/06/gut3.png) Geographic distribution of obesity related gut bacteria. Pie charts indicate the relative abundance of obesity-associated Firmicutes bacteria (blue) vs. Bacteroidetes bacteria (red) in 1,020 individuals. From Sanders R. (2013.) Berkeley News Center, http://newscenter.berkeley.edu/2014/02/14/geographic-variation-of-human-gut-microbes-tied-to-obesity/.[/caption]
Such a shift may have been an evolutionary adaptation to colder climates given Firmicutes’ enhanced ability to extract energy from food (7). As Suzuki stated in a recent interview(8), “People think that obesity is a bad thing, but maybe in the past getting more fat and more energy from the diet might have been important to survival in cold places. This suggests that what we call ‘healthy microbiota’ may differ in different geographic regions.” Suzuki’s advisor, Professor Michael Nachman, agrees. “Bergmann’s rule-that body size increases with latitude for many animals - is a good one and presumed to be an adaptation for dealing with cold environments,” Nachman said.
Age is another factor that can drastically impact gut microbiome composition. Changes in host behavior and diet as well as tissue structure and gene expression have all been shown to effect microbe type and abundance (1,4,13). Infants have higher levels of Lactobacillus bacteria for breaking down a milk-rich diet, bacterial composition in the adult stomach can vary according to the presence or absence of the ulcer associated H*. pylori* bacterium, and a decrease in overall microbial diversity is associated with the elderly.
Professor Jillian Banfield in the Department of Earth and Planetary Sciences at UC Berkeley investigates premature infant gut composition as a means of tracking some of the earliest microbial colonization events in humans. Until birth, the infant gut is considered sterile with delivery method (vaginal vs. Caesarean), feeding choice (bottle vs. breast feeding) and even environment (hospital vs. home) providing some of the earliest microbe exposures (4,10-12).
[caption id="attachment\\_12834" align="aligncenter" width="500"][](http://berkeleysciencereview.com/wp-content/uploads/2015/06/gut4.png) How infants acquire gut microbes. From Cho I, Blaser MJ. (2012).The human microbiome: at the interface of health and disease. Nat Rev Gen. 13(4), 260-270.[/caption]
Banfield’s time course analysis of infant fecal samples revealed that their gut undergoes several waves of colonization by bacteria and viruses during the first few weeks of life (10-12). Firmicutes and other fermentative bacteria overtake E. coli as the most abundant species while several skin associated bacteria persist at fairly consistent levels (10,11). Many microbes matched those found in the neonatal intensive care unit, providing further evidence for the role of environmental exposure in gut development (12). Banfield’s research team also refined microbiome identification techniques so that both species type and genetic variants or strains could be identified.
With such refined techniques, could we have access to our own individual gut fingerprints? Yes and No. Even with enhanced analysis such as those developed by Banfield, a substantial amount of microbiome associated DNA remains unidentified (1,4). Microbe detection and identification methods also vary extensively between research groups with the choice of sample source material (fecal vs. intestinal tissue), genes for identifying specific microbes, computational software, and even collection methods contributing to conflicting results (2). Until collection and classification methods are standardized, consensus on the “core microbiome” or a healthy gut profile shared between all individuals remains elusive. Still, with significant associations demonstrated between gut composition and diseases including obesity and cancer, the impact of our microbial communities on overall well-being cannot be ignored.
[caption id="attachment\\_12830" align="aligncenter" width="1024"][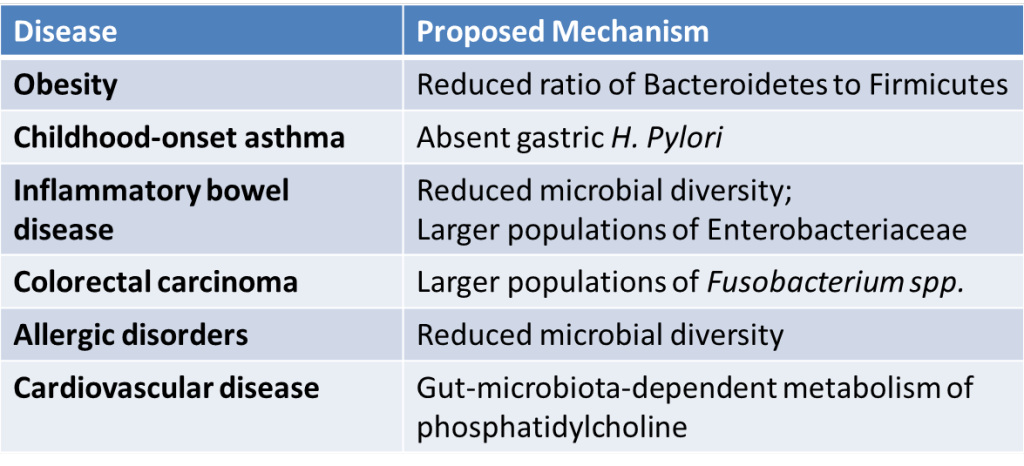](http://berkeleysciencereview.com/wp-content/uploads/2015/06/gut5.png) Diseases with potential links to the gut microbiome. Derived from Khanna S, Tosh P. (2014). Mayo Clin Proc. 89(1), 107-114. and Cho I, Blaser MJ. (2012). Nat Rev Gen. 13(4), 260-270.[/caption]
Guts guidance is already practiced on a daily basis. Doctors routinely treat excessive acid production by prescribing antibiotics against the ulcer associated H*. pylori* bacteria while the consumption of probiotics or live microorganisms is consistently used for the temporary relief of mild gastrointestinal symptoms (14). Knowing your own individual fingerprint in the future could lead to customized treatments that alter your gastrointestinal flora for everything from dieting to disease. Trying to lose weight? Take a Bacteroidetes booster. Trying to prevent food allergies? Increase your Clostridia levels to alter the sensitivity of your immune system (15). Gut fingerprinting could also prevent epidemics and reduce malnutrition. The identity of “super spreaders“ or the individuals responsible for the majority of disease transmissions could be determined by their microbiome profile of antibiotic resistance (16). Nutritional interventions that increase biodiversity in the gut flora of malnourished children also successfully diminished symptoms (16, 17). The national Human Microbiome Project and the European MetaHIT project continue to expand our knowledge of this diverse, unseen community through their sequence- specific data. So go ahead and listen to your guts guidance - you may be surprised where it leads you.
References
Qin J, Li R, Raes J, Manimozhiyan A, Burgdorf KS, Chaysavanh M et al (2010). A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 464(7285):59-65.
Tyler AD, Smith MI, Silverberg MS. (2014.) Analyzing the Human Microbiome: A “How To” guide for physicians. Am J Gastroenterol. 109(7), 983-993.
Degnan PH, Taga ME, Goodman AL. (2014.) Vitamin B12 as a Modulator of Gut Microbial Ecology. Cell Metabolism. 20(5), 769-778.
Cho I, Blaser MJ. (2012).The human microbiome: at the interface of health and disease. Nat Rev Gen. 13(4), 260-270.
Franzosa EA, Huang K, Meadow JF, Gevers D, Lemon KP, Bohannan BJ, Huttenhower C. (2015.) Identifying personal microbiomes using metagenomic codes. Proc Natl Acad Sci USA. www.pnas.org/cgi/doi/10.1073/pnas.1423854112.
Greenblum S, Turnbaugh PJ, Borenstein E. (2012.) Metagenomic systems biology of the human gut microbiome reveals topological shifts associated with obesity and inflammatory bowel disease. Proc Natl Acad Sci USA. 109(2), 594-599.
Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. (2006). An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 444 (21/28), 1027-1031.
Suzuki TA, Worobey M. (2014.) Geographical variation of human gut microbial composition. Biol Lett. 10: 20131037. http://dx.doi.org/10.1098/rsbl.2013.1037.
Sanders R. (2013.) Geographic variation of human gut microbes tied to obesity. Berkeley News Center, http://newscenter.berkeley.edu/2014/02/14/geographic-variation-of-human-gut- microbes-tied-to-obesity/.
Sharon I, Morowitz MJ, Thomas BC, Costello EK, Relman DA, Banfield JF. (2013, Jan). Time series community genomics analysis reveals rapid shifts in bacterial species, strains and phage during infant gut colonization. Genome Res. 23(1), 111-120.
Brown CT, Sharon I, Thomas BC, Castelle CJ, Morowitz MJ, Banfield JF.(2013, Dec). Genome resolved analysis of premature infant gut microbial community reveals a Varibaculum cambriense genome and a shift towards fermentation-based metabolism during the third week of life. Microbiome. 1(1), doi: 10.1186/2049-2618-1-30.
Brooks B, Firek BA, Miller CS, Sharon I, Thomas BC, Baker R, Morowitz MJ, Banfield JF. (2014). Microbes in the neonatal intensive care unit resemble those found in the gut of premature infants. Microbiome. 2(1), doi: 10.1186/2049-2618-2-1.
Khanna S, Tosh P. (2014). A Clinician’s Primer on the Role of the Microbiome in Human Health and Disease. Mayo Clin Proc. 89(1), 107-114.
Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, et al. (2014). Expert consensus document.The International Scientific Association for Probiotics and Prebiotics consensusstatement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol.11(8), 506-514.
Stefka AT, Feehley T, Tripathi P, Qiu J, McCoy K, Mazmanian SK, et al. (2014). Commensal bacteria protect against food allergen sensitization. Proc Natl Acad Sci USA. 111(36), 13145- 13150.
Gopinath S, Lichtman JS, Bouley DM, Elias JE, Monack DM. (2014). Role of disease- associated tolerance in infectious superspreaders. Proc Natl Acad Sci USA. 111(44), 15780-15785.
Subramanian S, Huq S, Yatsunenko T, Haque R, Mahfuz M, Alam MA, et al. (2014). Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature. 510(7505), 417-421.



